Abstract
Introduction: Despite recent advances in treatment for acute myeloid leukemia (AML), AML remains a deadly disease, with approximately 70% of patients succumbing to the disease within five years of diagnosis (SEER database 2021). Intensive induction chemotherapy carries a risk of morbidity and mortality, making it a risky treatment for patients who are unlikely to achieve a remission with DNA damaging-based treatments. Hypomethylating agents (HMAs) such as azacitidine (AZA) and decitabine (DAC) are now frequently used in AML treatment as less intensive treatment options. Thus, biomarkers to predict response to DNA damaging agents in AML are needed. We recently found a significant association between AML patient survival and high expression of Schlafen11 (SLFN11), a dynamically inducible key effector of the DNA replication stress response. SLFN11 has been shown to be associated with response to certain chemotherapy agents in some solid tumors in vitro and in vivo (Conteduca et. al. JCO 2019; Shee et. al. PLoS One 2019), but its importance in leukemia has not been well defined. It is also unknown whether SLFN11 levels affect response to HMAs. We hypothesized that SLFN11 can be used as a predictive biomarker for treatment response to DNA-damaging agents in AML but that response to HMAs would be independent of HMAs. We also hypothesized that identifying pathways upregulated in SLFN11-low cells could lead to novel combination therapies to increase sensitivity to chemotherapeutics.
Methods: The human leukemia cell lines HEL and U937 cells were used for these studies. U937- and HEL-SLFN11 knockout (SLFN11KO) cells were generated using CRISPR-Cas9 gene editing technology. Cell viability was assessed by WST-1 reagent assay. SLFN11 and other protein levels were measured by Western blotting.
Results: SLFN11 protein expression was readily observed by immunoblotting of HEL and U937 cell lysates. U937- and HEL-SLFN11KO cells were incubated with increasing concentrations of cytarabine (AraC), which is the backbone of most chemotherapy regimens currently used in AML treatment. Both U937- and HEL-SLFN11KO cells were more resistant to AraC than their respective control cells. To examine the mechanism of resistance in U937-SLFN11KO cells, SLFN11KO and control cells were treated with AraC for up to 8 hrs, and lysates were subjected to immunoblotting for various DNA damage response proteins. There were higher baseline levels of multiple DNA damage-related proteins (including Chk1 and Chk2) in the SLFN11KO cells. There was also prolonged activation of the DNA damage pathway in U937-SLFN11KO cells, as evidenced by phosphorylation of the ATR substrate Chk1 (Ser345) through 8 hours, whereas phospho-Chk1 levels decreased in control cells by 8 hours. Interestingly, this prolonged activation occurred without a concomitant increase in γH2AX (a marker of DNA damage and double strand breaks).
Because U937-SLFN11KO cells demonstrated a more pronounced activation of the ATR/Chk1 pathway in response to AraC, we subjected U937-SLFN11KO and control cells to increasing concentrations of AraC in combination with a small dose (0.3 μM) of the ATR inhibitor VE822. Notably, ATR inhibition restored sensitivity of SLFN11KO cells to AraC. These results suggest that SLFN11KO cells are resistant to DNA damaging agents via upregulation and activation of DNA repair proteins, and that adding inhibitors of these proteins can decrease resistance to chemotherapy agents in SLFN11KO cells. In contrast, SLFN11KO cells remained sensitive to AZA and DAC, suggesting that HMAs may be preferable to AraC for patients with SLFN11-low AML.
Conclusions: Low SLFN11 levels predict poor response to standard chemotherapy agents (i.e. AraC) in AML cells, suggesting that SLFN11 may prove to be an important predictive biomarker for AML patients. In addition, SLFN11-low AML cells have an increased reliance on DNA repair pathways, and inhibition of DNA repair proteins such as ATR may enhance responses to AraC in these cells. This raises the possibility of patient selection based on SLFN11 levels for specific combination approaches for the treatment of AML.
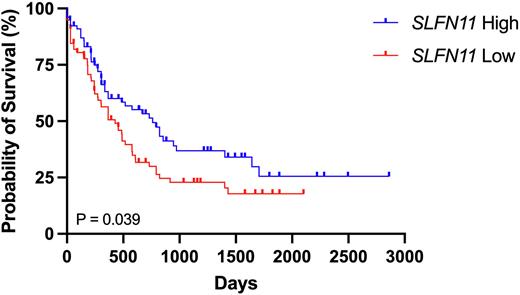
Figure 1. Kaplan Meier survival curves of AML patients in the TCGA (LAML) database with high (top 50th percentile) or low (bottom 50th percentile) SLFN11 expression using UCSC Xena (Goldman et. al. Nat Biotechnol 2020).
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal